Convert a volumetric file (.vol, .longvol, .pgm3d) into a set of 2D slice images.
Usage: vol2slice [input] [output]
Allowed options are:
-h [ --help ] display this message
-i [ --input ] arg vol file (.vol, .longvol .p3d, .pgm3d and
if WITH_ITK is selected: dicom, dcm, mha,
mhd). For longvol, dicom, dcm, mha or mhd
formats, the input values are linearly
scaled between 0 and 255.
-o [ --output ] arg base_name.extension: extracted 2D slice
volumetric files (will result n files
base_name_xxx.extension)
-f [ --setFirstSlice ] arg (=0) Set the first slice index to be extracted.
-l [ --setLastSlice ] arg Set the last slice index to be extracted
(by default set to maximal value according
to the given volume).
-s [ --sliceOrientation ] arg (=2) specify the slice orientation for which
the slice are defined (by default =2 (Z
direction))
--rescaleInputMin arg (=0) min value used to rescale the input
intensity (to avoid basic cast into 8
bits image).
--rescaleInputMax arg (=255) max value used to rescale the input
intensity (to avoid basic cast into 8 bits
image).
Example:
# Export Z slice images (-s 2):
$ vol2slice -i ${DGtal}/examples/samples/lobster.vol -o slice.pgm -f 10 -l 15 -s 2
You should obtain such a visualization:
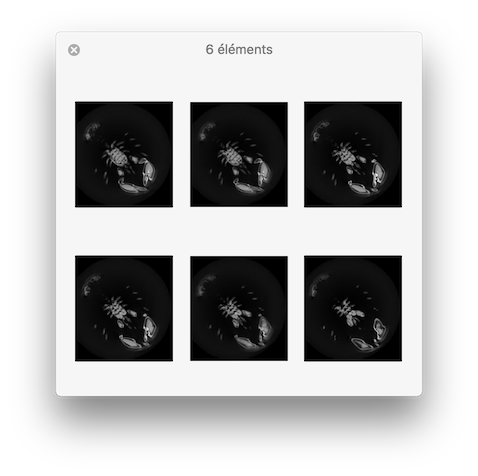
resulting visualisation.
- See also
- vol2slice.cpp

 1.8.10
1.8.10