Marching-cube like surface extracted using the combinatorial manifold structure of digital surfaces.
* # Commands * $ ./examples/topology/volMarchingCubes ../examples/samples/Al.100.vol 0 1 0 *
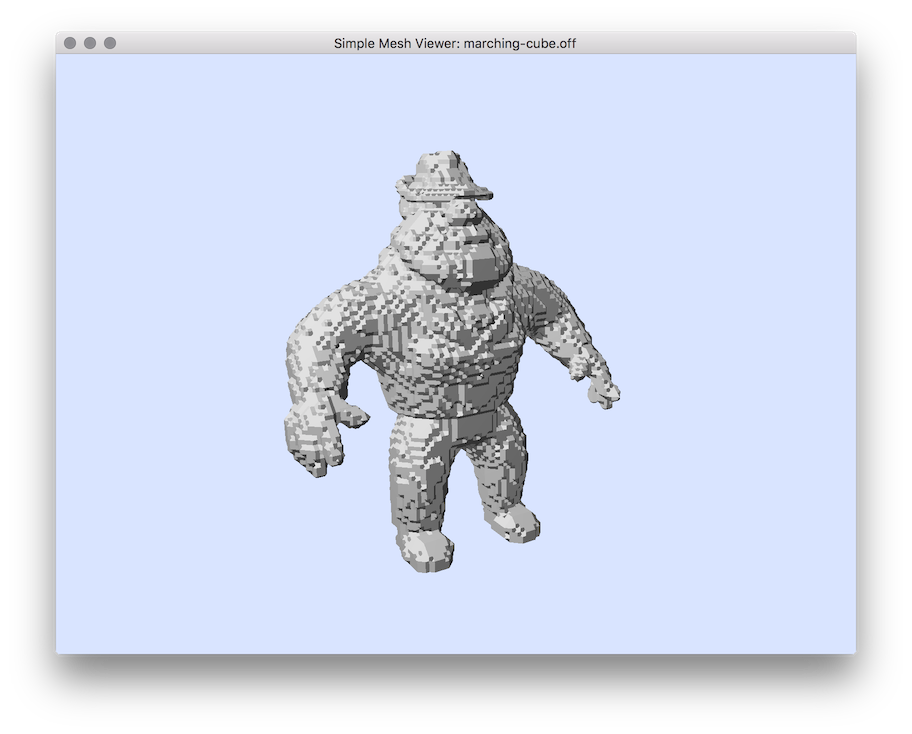
Marching-cube surface of Al.100.vol file.
#include <iostream>
#include <queue>
#include "DGtal/base/Common.h"
#include "DGtal/kernel/CanonicEmbedder.h"
#include "DGtal/io/readers/VolReader.h"
#include "DGtal/images/ImageSelector.h"
#include "DGtal/images/imagesSetsUtils/SetFromImage.h"
#include "DGtal/images/ImageLinearCellEmbedder.h"
#include "DGtal/shapes/Shapes.h"
#include "DGtal/helpers/StdDefs.h"
#include "DGtal/topology/helpers/Surfaces.h"
#include "DGtal/topology/DigitalSurface.h"
#include "DGtal/topology/SetOfSurfels.h"
using namespace std;
using namespace DGtal;
using namespace Z3i;
{
std::cerr << "Usage: " << argv[ 0 ] << " <fileName.vol> <minT> <maxT> <Adj>" << std::endl;
std::cerr << "\t - displays the boundary of the shape stored in vol file <fileName.vol>." << std::endl;
std::cerr << "\t - voxel v belongs to the shape iff its value I(v) follows minT <= I(v) <= maxT." << std::endl;
std::cerr << "\t - 0: interior adjacency, 1: exterior adjacency." << std::endl;
}
{
if ( argc < 5 )
{
usage( argc, argv );
return 1;
}
std::string inputFilename = argv[ 1 ];
unsigned int minThreshold = atoi( argv[ 2 ] );
unsigned int maxThreshold = atoi( argv[ 3 ] );
bool intAdjacency = atoi( argv[ 4 ] ) == 0;
Image image = VolReader<Image>::importVol(inputFilename);
DigitalSet set3d (image.domain());
SetFromImage<DigitalSet>::append<Image>(set3d, image,
minThreshold, maxThreshold);
KSpace ks;
bool space_ok = ks.init( image.domain().lowerBound(),
image.domain().upperBound(), true );
if (!space_ok)
{
return 2;
}
typedef SurfelAdjacency<KSpace::dimension> MySurfelAdjacency;
MySurfelAdjacency surfAdj( intAdjacency ); // interior in all directions.
typedef SetOfSurfels< KSpace, SurfelSet > MySetOfSurfels;
MySetOfSurfels theSetOfSurfels( ks, surfAdj );
Surfaces<KSpace>::sMakeBoundary( theSetOfSurfels.surfelSet(),
ks, set3d,
image.domain().lowerBound(),
image.domain().upperBound() );
MyDigitalSurface digSurf( theSetOfSurfels );
<< std::endl;
typedef CanonicEmbedder< Space > MyEmbedder;
typedef
ImageLinearCellEmbedder< KSpace, Image, MyEmbedder > CellEmbedder;
CellEmbedder cellEmbedder;
MyEmbedder trivialEmbedder;
// The +0.5 is to avoid isosurface going exactly through a voxel
// center, especially for binary volumes.
cellEmbedder.init( ks, image, trivialEmbedder,
( (double) minThreshold ) + 0.5 );
ofstream out( "marching-cube.off" );
if ( out.good() )
digSurf.exportEmbeddedSurfaceAs3DOFF( out, cellEmbedder );
out.close();
return 0;
}
std::ostream & error()
void beginBlock(const std::string &keyword="")
std::ostream & info()
double endBlock()
DigitalSurface< MyDigitalSurfaceContainer > MyDigitalSurface
Definition: greedy-plane-segmentation-ex2.cpp:92
DigitalSetSelector< Domain, BIG_DS+HIGH_BEL_DS >::Type DigitalSet
Definition: StdDefs.h:100
DGtal is the top-level namespace which contains all DGtal functions and types.
Definition: ClosedIntegerHalfPlane.h:49
ImageContainerBySTLVector< Domain, Value > Image
Definition: testSimpleRandomAccessRangeFromPoint.cpp:45