This part of the manual describes how to import and export images and main DGtal objects from/to various formats.
Introduction
In DGtal, file readers and writers are located in the "io/readers/" and "io/writers/" folders respectively. Most of them are dedicated to image format import/export but some other DGtal data structures can have such tools (e.g. point set/mesh readers).
Before going into details, let us first present an interesting tool for image visualisation or image export: predefined colormaps to convert scalars or to (red,green,blue) triplets.
Colormaps
Colormap models satisfy the CColormap concept. For short, a colormap is parametrized by a scalar value template type (Value). When constructed from two min and max values (of type Value), the colormap offers an operator returning a DGtal::Color for each value v in the interval [min,max].
For example, RandomColorMap returns a random color for each value v. More complex colormaps (GradientColorMap, HueShadeColorMap, ...) offer better colormap for scientific visualisation purposes.
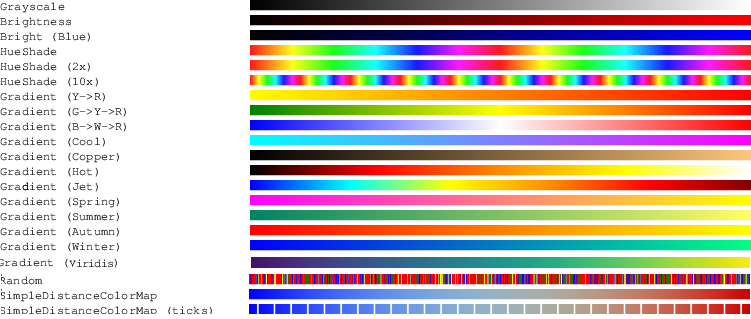
Beside colormaps, TickedColorMap is a colormap adapter that adds ticks. For example, you can adapt a Black-Red gradient colormap to add regular white ticks (see usage in testTickedColorMap.cpp):

Ticks can be regularly spaced or explicitly given by the user.
In some situations, we may have to convert colors into scalar values (see below). In this case, basic conversion functors are available in the DGtal::functors namespace. For example, you would find in this namespace a DGtal::functors::RedChannel converter or a DGtal::functors::MeanChannels converter.
Hence, to implement a functor taking values and returning the red channel of a colormap, you just have to compose the two functors with the help of the Composer:
Image file formats
We first detail import/export format for DGtal images. Please refer to moduleImages for details on images in DGtal and their associated concepts. First of all:
- Image readers are classes templated by an image container type.
- Image writers are classes templated by an image type and a functor to convert image values to the value type of the writer file format.
Hence, for image writers, some functors may return a DGtal::Color or a scalar value depending on the writer.
Image Writers
| Dimension | Name | Description | Functor requirements | Class name | Comment |
|---|---|---|---|---|---|
| 2D | PGM | Grayscale netpbm image format | the functor should return an unsigned char | PGMWriter | http://netpbm.sourceforge.net/ |
| PPM | Color netpbm image format | the functor should return a DGtal::Color | PPMWriter | http://netpbm.sourceforge.net/ | |
| any stb_image format | Color image format (png,jpg, bmp,tga ) | the functor must return a DGtal::Color | STBWriter | ||
| any 2D ITK format | Grayscale or 2D ITK image | the functor should return a ITKIOTrait<Image::Value>::ValueOut | ITKWriter | with DGTAL_WITH_ITK build flag, http://www.itk.org/ | |
| 3D | PGM3D | Grayscale netpbm image format | the functor should return an unsigned char | PGMWriter | |
| PPM3D | Color netpbm image format | the functor should return a DGtal::Color | PPMWriter | ||
| Vol | Volumetric file format | the functor should return an unsigned char | VolWriter | Simplevol project, http://liris.cnrs.fr/david.coeurjolly | |
| Longvol | Volumetric file format (long) | the functor should return a DGtal::uint64_t | LongvolWriter | Simplevol project, http://liris.cnrs.fr/david.coeurjolly | |
| HDF5 | HDF5 file with 3D UInt8 image dataset(s) | the functor should return a DGtal::uint8_t | HDF5Writer | with DGTAL_WITH_HDF5 build flag, http://www.hdfgroup.org/HDF5/ | |
| any 3D ITK format | Any 3D ITK image | the functor should return a ITKIOTrait<Image::Value>::ValueOut | ITKWriter | with DGTAL_WITH_ITK build flag, http://www.itk.org/ | |
| nD | Raw8 | raw binary file format on 8bits | the functor should return an unsigned char | RawWriter | |
| Raw16 | raw binary file format on 16bits | the functor should return an unsigned short | RawWriter | ||
| Raw32 | raw binary file format on 32bits | the functor should return an unsigned int | RawWriter | ||
| Raw | raw binary file format for any type | the functor should return the same type as specified in the template parameter of RawWriter::exportRaw | RawWriter |
- Note
- Since DGtal doesn't integrate ITK by default, ITK image should by writen directly using the ITKWriter class. ImageContainerBySTLVector< Domain, Value > ImageDefinition testSimpleRandomAccessRangeFromPoint.cpp:45
For scalar value format (PGM, Vol, Longvol, Raw, ...), the associated template class have a default functor type. Hence, if you just want to cast your image values to the file format value type (e.g. "unsigned char" for Vol), do not specify any functor.
The class GenericWriter allows to automatically export any image (2d, 3d, nd) from its filename. The class is templated with an image container type, a dimension value (given by default by the image container dimension), a value type, (also given by default by the image container) and a functor type (by default set to the DefaultFunctor type).
To use it you need first to include the following header:
After constructing and filling an image (anImage2D or anImage3D), by default you can save it with:
As the other export functions, a functor can be used as optional argument (as given in the previous example):
If you don't need to specify special functor and if don't need to change default image type, you can use a less generic writer with the stream operator and the string filename:
- Note
- Naturally, the stream operator hide a default functor (c++ cast to a given type).
To write color images, you need to use a functor which transform a scalar value into a Color. You can use the previous Colormaps :
Image Readers
| Dimension | Name | Description | Class name | Comment |
|---|---|---|---|---|
| 2D | PGM | Grayscale netpbm image format | PGMReader | http://netpbm.sourceforge.net/ |
| any stb format | Any file format in the stb_image library (bmp,png,jpg,tga, gif) | STBReader | ||
| HDF5 | HDF5 file with 2D image dataset(s) | HDF5Reader | with DGTAL_WITH_HDF5 build flag, http://www.hdfgroup.org/HDF5/ | |
| any 2D ITK format | Any file format in the ITK library (png, jpg, mhd, mha, ...) | ITKReader | with DGTAL_WITH_ITK build flag, http://www.itk.org/ | |
| 3D | PGM3D | Grayscale netpbm image format | PGMReader | |
| PGM | Same as PGM3D but with same header as in 2D (P2 or P5) | PGMReader | ||
| DICOM | Medical format (from scanners, with the use of ITK library) | DicomReader | http://medical.nema.org/ | |
| Vol | Volumetric file format | VolReader | Simplevol project, http://liris.cnrs.fr/david.coeurjolly | |
| Longvol | Volumetric file format (long) | LongvolReader | Simplevol project, http://liris.cnrs.fr/david.coeurjolly | |
| HDF5 | HDF5 file with 3D UInt8 image dataset(s) | HDF5Reader | with DGTAL_WITH_HDF5 build flag, http://www.hdfgroup.org/HDF5/ | |
| any 3D ITK format | Any file format in the ITK library (mhd, mha, ...) | ITKReader | with DGTAL_WITH_ITK build flag, http://www.itk.org/ | |
| nD | Raw8 | raw binary file format on 8bits | RawReader | |
| Raw16 | raw binary file format on 16bits | RawReader | ||
| Raw32 | raw binary file format on 32bits | RawReader | ||
| Raw | raw binary file format for any type | RawReader |
- Note
- Since DGtal doesn't integrate ITK by default, ITK image should by read directly using the ITKReader class. static Image importITK(const std::string &filename, const TFunctor &aFunctor=TFunctor(), bool shiftDomainUsingOrigin=true)
The class GenericReader allows to automatically import any image (2d, 3d, nd) from its filename. The class is templated with an image container type, a dimension value (given by default by the image container dimension), a value type, (also given by default by the image container). Note that the reader choice between 8 bits or 32 bits is automatically done according to the templated image container type. So you can have an DGtal::IOException if you try to read a 8 bits raw image from an unsigned int image type (choose an 8 bits type like unsigned char or explicitly call the specific reader RawReader::importRaw8).
Use the same import function for both 2D or 3D images:
Details on the Vol/Longvol formats
The Vol and Longvol formats are described on the simplevol project website (http://liris.cnrs.fr/david.coeurjolly/code/simplevol.html). Basically, the format consists in:
- an ASCII header with for instance the image size and the Vol format version, ending with a ".". Mandatory information are the size (X, Y, Z) and the format version (Version). E.g. Center-X: 5Center-Y: 5Center-Z: 5X: 11Y: 11Z: 11Voxel-Size: 1Alpha-Color: 0Voxel-Endian: 0Int-Endian: 0123Version: 3.
- a binary chunck with linearized image values. In "Version 2" of Vol and Longvol, the binary chunck is simply a raw export of image values (little endian for Longvol entries). In "Version 3", the binary values are compressed using zlib (default version when using the DGtal reader and writer).
If you want to export an image to "Version 2" Vol or a Longvol, just add a false flag when exporting. For instance:
- Note
- "Version 1" Vol or Longvol files are no longer supported in DGtal readers/writers.
Other geometrical formats
Point list format in n-D
The static class PointListReader allows to read discrete points represented in simple file where each line represent a single point.
3D Surface Mesh
The static class MeshReader allows to import Mesh from OBJ, OFF or OFS file format. Actually this class can import surface mesh (Mesh) where faces are potentially represented by triangles, quadrilaters and polygons. Notes that Mesh can be directly displayed with PolyscopeViewer.
The mesh importation can be done automatically from the extension file name by using the "<<" operator. For instance (see. Import 3D mesh from OFF file ):
You can also export a Mesh object by using the operator (">>").
Sparse Voxel Octree and Sparse Voxel Direct Acyclic Graphs
The static classes SVOReader and SVOWriter allow to import and export DigitalSetByOctree from and to files. In order to maintain an efficient data representation, DGtal uses a custom file format described below. Both SVO and SVDag are stored using the same file fomat.
Theses classes can be used as follows to import or export SVO:
The reader allows an octree to be read in a different space of the same dimension. However, there may be integer truncation that may lead to unexpected results (a warning is issued in this case). Please note that BigIntegers are not supported for reading or writing.
SVO File format
The SVO file format is comprised of a header in ascii, and data in binary.
The header is written in ascii. Each line represents a different fields, whose name is space and case sensitive and for which the value is separated by a ':'. The value is written in ASCII format, and surrounding whitespace (spaces, tabs, carriage returns) is trimmed. There is no specified order, but we recommand following the order bellow. The header ends with a single '.' on a separated line. The expected fields are:
- 'Format': always 'SVO'.
- 'Version': unsigned integer, for now only '1' is supported.
- 'Size': unsigned integer representing the number of voxels stored.
- 'Dim': unsigned integer representing the dimension of voxels.
- 'LowerBound': exactly 'Dim' integers separated by spaces representing the domain lower bound.
- 'UpperBound': exactly 'Dim' integers separated by spaces representing the domain upper bound.
- 'State': Indicates whether this is an SVO ('1') or an SVDag ('2').
- 'Compression': Indicates whether data is compressed ('1') or not ('0').
For example, a header could be:
The data is always written in binary in little-endian order (regardless of host system). When compressed, data is compressed using zlib algorithm with the best compression level (9). The data is layed out as follows:
- 8 bytes: 'SizeSize', the number of bytes array length occupies in memory
- 8 bytes: 'PtrSize', the number of bytes array indices occupies in memory
- [SizeSize] bytes: 'Depth', the depth of the underlying tree.
- [Depth] * [SizeSize] bytes: 'ChildrenCount', the number of children at each depth of the tree. /bin/bash: line 1: :w: command not found
Examples
Importing a 3D digital set from volume file:
Importing and visualizing a digital set from a vol file can be done in few code lines. (see. digitalSetFromVol.cpp).
First we select the Image type with int:
Then the initial image is imported:
Afterwards the set is thresholded in ]0,255[:
Then you will obtain the following visualisation:

Import 3D point list from file
The example digitalSetFromPointList.cpp shows a simple example of 3d set importation:
We can change the way to select the coordinate field:
You may obtain the following visualisation:
Import 3D mesh from OFF file
The following example meshFromOFF.cpp shows in few lines how to import and display an OFF 3D mesh. Add the following headers to access to OFF reader and PolyscopeViewer:
then import an example ".off" file from the example/sample directory:
Display the result:
You may obtain the following visualisation:

You can also import large scale mesh, like the one of classic Angel scan ( available here: http://www.cc.gatech.edu/projects/large_models/ )
