volSegment
Segments volumetric file from a simple threshold which can be set automatically from the otsu estimation.
Usage: volSegment [input] [output]
Allowed options are :
Positionals:
1 TEXT:FILE REQUIRED volumetric input file (.vol, .pgm, .pgm3d, .longvol)
2 TEXT:FILE REQUIRED volumetric output file (.vol, .pgm, .pgm3d, .longvol)
Options:
-h,--help Print this help message and exit.
-i,--input TEXT:FILE REQUIRED volumetric input file (.vol, .pgm, .pgm3d, .longvol)
-o,--output TEXT=result.vol volumetric output file (.vol, .pgm, .pgm3d, .longvol)
--labelBackground option to define a label to regions associated to object background.
-m,--thresholdMin INT=0 min threshold (if not given the max threshold is computed with Otsu algorithm).
-M,--thresholdMax INT=255 max threshold
Example:
You can test the segmentation in the lobster volume file:
$ volSegment ${DGtal}/examples/samples/lobster.vol segmentation.vol -m 70 -M 255
Definition: ATu0v1.h:57
You will obtain a volumetric file representing for each voxel a label associated to a connected component. You can display this segmentation results by extracting it in SDP format with the vol2sdp tool (with option -e to export also the image labels):
$ vol2sdp segmentation.vol segmentation.sdp -e -m 1 -M 255
and display them with 3DSDPViewer :
$ 3dSDPViewer segmentation.sdp --importColorLabels
You should obtain such a result:
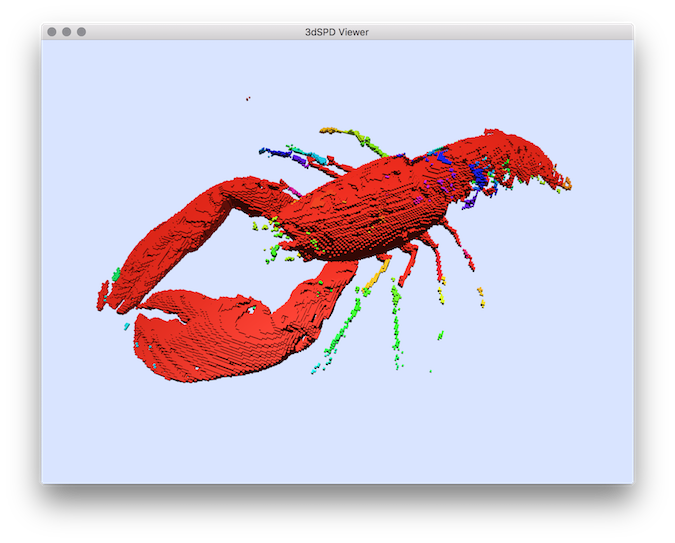
Segmentation result displayed with colors representing the segmentation labels.
- See also
- volSegment.cpp
 1.9.1
1.9.1